Available online at: https://doi.org/10.18778/1898-6773.87.3.01
 https://orcid.org/0000-0002-6302-8698
https://orcid.org/0000-0002-6302-8698
Department of Anthropology, Burdur Mehmet Akif Ersoy University, Burdur, Turkey
 https://orcid.org/0000-0001-9792-1508
https://orcid.org/0000-0001-9792-1508
Department of Anthropology, Binghamton University, Binghamton, New York, United States
ABSTRACT: Shovel-shaped incisors (SSI) and Carabelli’s cusp (CC) are noteworthy human dental non-metric traits which presence and degree of expression have been reported to cluster within distinct populations. Recent advances in developmental biology suggest that SSI and CC are likely under polygenic developmental control; therefore, genetic variation in multiple genes is likely to contribute to differential SSI and CC expression. The exact genetic mechanisms underlying variation in SSI and CC development, however, remain mostly unknown. This study aims to identify whether variation in the basal DNA sequences of six candidate genes, NKX2-3, SOSTDC1, BMP4, FGF3, FGF4, and WNT10A, has any impact on SSI and/or CC expression. Study methods involve collection of saliva samples and dental data from 36 participants. The Arizona State University Dental Anthropology System (ASUDAS) has been used to score SSI and CC expression. Next-generation sequencing (NGS) methods were utilized to sequence the entire gene region of the candidate genes. Spearman’s correlation test was used to score the relationship between the genotypeand degree of trait expression of participants. Fifteen SNPs/INDELs belonging to SOSTDC1, FGF3, FGF4 and WNT10A were significantly associated with SSI and/or CC expression. No SNPs/INDELs were detected in the genes BMP4 and NKX2-3 that significantly contributes to observed phenotypes. FGF3, FGF4, SOSTDC1 and WNT10A were possibly involved in the formation of shoveling and Carabelli’s cusp. However, because of the small sample size, more studies are needed to confirm their role and rule out any potential role of NKX2-3 and BMP4 in the production of SSI and CC.
KEY WORDS: dental nonmetric traits, Next Generation Sequencing, long-range PCR, variant identification, genotype-phenotype
Teeth develop through acomplex and multidimensional process called odontogenesis, which involves several evolutionarily conservative elements interacting during embryonic and postnatal development. Genetic, epigenetic, and environmental factors are the main factors that can alter the final dental morphology (Brook et al. 2014). Therefore, the normal dental variation we observe within apopulation is the result of polygenic and environmental influences working in concert. Due to the multifactorial nature of tooth development, identification of the causal biological mechanisms that result in dental characteristics is rather challenging (Scott et al. 2018).
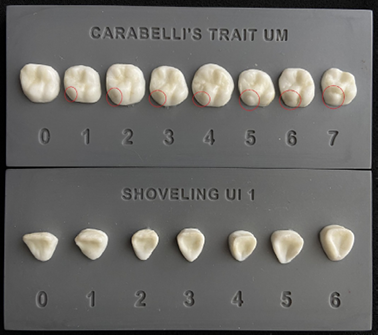
Nonmetric dental traits are inheritable features of teeth that exhibit patterned geographic variation due to the strong genetic component involved during their development (Scott et al. 2018). Previous work posited that the nonmetric dental traits are selectively neutral and thus do not easily adapt to the changing environmental conditions (Irish et al. 2020). Therefore, anthropologists have used nonmetric dental traits as proxies for genetic relatedness within and between human populations (Hanihara 2008; Scott et al. 2018). Currently, over 40 dental variants have been described by the ASUDAS (Arizona State University Dental Anthropology System) (Turner et al. 1991; Scott and Irish 2017). Among these variants, shovel-shaped incisors (SSI) and Carabelli’s cusp (CC), which are the focus of this study, arethe two dental features that have drawn thegreatest attention from anthropologists (Fig. 1)(Carayon et al. 2019). SSI is prevalent, with the occurrence nearing 100%, in some North and East Asian, as well as Native American groups (Hlusko et al. 2018; Scott et al. 2018). On the other hand, populations with apredominance of European ancestry frequently exhibit CC (Scott and Irish 2017). SSI and CC have awell-established global distribution (Hrdlička 1920; Scott 1980; Tsai et al. 1996; Scott et al. 2018).
During development, reciprocal interactions between chemical signals within and around the cells of adeveloping tooth determine the final dental phenotype (Hughes and Townsend 2013). Two tissue layers involved in tooth development, the dental epithelium and the underlying mesenchyme, play intricate roles in gene activation and silencing which results in the development of the dental nonmetric characteristics (Jernvall and Thesleff 2000; Thesleff 2006; Jussila and Thesleff 2012; Scott et al. 2018). Currently, over 300 genes have been identified that are involved in the process of odontogenesis (Thesleff 2006). Still, the precise genetic pathways influencing the development of distinct nonmetric dental features are not fully understood. Further research is needed to identify genes involved in the formation of dental nonmetric traits.
This study aims to provide an initial investigative framework for understanding the genetic and genomic regulatory mechanisms that induce the production of two anthropologically and forensically relevant discrete dental traits: shovel-shaped incisors and Carabelli’s cusp. We employed the candidate gene approach, which required the selection of the pre-specified genes of interest that were previously demonstrated to be involved in tooth development (Tabor etal. 2002). This study aims to determine whether the polymorphisms in NKX2-3, FGF3, FGF4, WNT10A, BMP4, and SOSTDC1 contributes to SSI and CC expression observed in an individual in order to better understand the genetic basis of geographic differences in tooth morphology.
The original objective of this project was to obtain dental impressions from the participants. However, due to the COVID-19 restrictions, we had to transition to aremote dental photo collection method instead of physical dental impressions. Individuals who showed interest in volunteering in the study received an informed consent form along with instructions for how to effectively and safely take dental photos. Participants were requested to take detailed photographs of their upper and lower teeth using adequate lighting to ensure clear visibility of the incisors and molars, which exhibit the two dental traits of interest in this study. Participants then emailed the photos to the first author of this study (i.e., FNE) to determine their eligibility to participate in the study. We also used the photos to score the dental traits of interest.
After the COVID-19 restrictions were lifted, in addition to dental pictures, we took negative impressions of upper and lower teeth from seventeen participants in order to make dental casts. Impressions were taken using vinyl polysiloxane (VPS), an alginate substitute dental impression material. The Arizona State University Dental Anthropology System (ASUDAS) plaques were used to score the dental traits (Figure 1). Only the incisors and molars on the left side of the dental arch was scored.
Saliva was collected using amouthwash collection protocol. The participants rinsed their mouths for 30 seconds with 10 ml of mouthwash before spitting into acollecting tube. DNA was extracted using the QIAamp DNA Mini Kit following the manufacturer’s instructions (Qiagen, Germany).
The National Center for Biotechnology Information (NCBI)’s Primer-BLAST tool was utilized to design long-range PCR primers with high specificity (Ye et al. 2012). The entire gene region of NKX2-3, FGF3, FGF4, BMP4, and SOSTDC1, including the intronic and exonic regions, were targeted in the primer design. WNT10A was separated into three smaller sections due to its length, and separate primer sets were designed for each section. However, only the last section of WNT10A was included in this study due to the unreliable amplification issues encountered for the first two sections. The primer sequences, genomic positions, melting temperatures (Tm) of the primer pairs, and amplicon lengths are displayed in Table 1.
The GeneAmp® PCR System 9700 (Applied Biosystems, Foster City, CA) was utilized to perform the PCRs. TaKaRa LA (Long and Accurate) Taq polymerase enzyme (Takara Bio Inc.) and reagents were used to amplify each gene area in 50 μl volumes. The entire gene area for NKX2-3, FGF3, FGF4, BMP4, and SOSTDC1 was amplified using atwo-step PCR method, which combines the annealing and extension stages. WNT10A was amplified utilizing the touch-down (TD) PCR method due to the specificity problems that we encountered. The PCR cycle conditions applied for each target region are displayed in Table 2. The 0.4% agarose gel electrophoresis was used to confirm the success of the PCR amplifications.
| Gene name | Genomic location (GRCh38/hg38) | Sequence (5’ → 3’) | Tm °C (Melting Temperature) | Length of amplicon (bp) |
| FGF3 | chr11:69809200-69820343 | Forward: CCAAGTGCCAGGAGAGGTTAGTACACTGC | 68.22 | 11144 |
| Reverse: GGGACAGAGGACCAGGAAGCAAGAGAAA | 67.64 | |||
| FGF4 | chr11:69770778-69776854& | Forward: TACAGTGCGGGAATGGCGTGAATTAGC | 67.46 | 6077 |
| Reverse: AGACAACACAGCAAGTGAGGGATGGGT | 67.87 | |||
| BMP4 | chr14:53949462-53959648 | Forward: CATCCCAGTGTTTCTCCAAGGCATGTGT | 67.17 | 10187 |
| Reverse: GGGCAGGACCAGGAAGTCTGCATTTCATC | 68.95 | |||
| NKX2-3 | chr10:99532030-99537060 | Forward: TTTGCCTCATTCAACCCTAGCAACAACCA | 67.10 | 5031 |
| Reverse: CTCCGCAAGTGACAAGGAGCCGCATA | 68.86 | |||
| SOSTDC1 | chr7:16458386-16465969 | Forward: TCTCACACCGAGCATCCTAAGTCACCTC | 67.23 | 7584 |
| Reverse: GCGTCGGCTCACAGACAAGTGATGAAGT | 68.79 | |||
| WNT10A | chr2:218886144-218894785 | Forward: TGTACCCAGAGAGGTGAGCTGGTGCAA | 69.04 | 8642 |
| Reverse: CACAAGAGGCCCAGGAAGAATGTGCCC | 69.30 | |||
| Total amplicon length | 48,665 |
| Target name | Cycle temperatures and times | Cycle Number | Cycle name |
| FGF3 | 94 °C 1 min | 1× | Initial denaturation |
| 98 °C 10 sec + 68 °C 10 min | 33× | Amplification | |
| 72 °C 10 min | 1× | Final elongation | |
| WNT10A | 94 °C 1 min | 1× | Initial denaturation |
| 98 °C 20 sec + 73 °C 5 min 40 sec | 5× | Amplification | |
| 98 °C 20 sec + 71 °C 5 min 40 sec | 5× | Amplification | |
| 98 °C 20 sec + 69 °C 5 min 40 sec | 25× | Amplification | |
| 72 °C 10 min | 1× | Final elongation | |
| NKX2-3 and FGF4 | 94 °C 1 min | 1× | Initial denaturation |
| 98 °C 10 sec + 69 °C 4 min 33 sec | 30× | Amplification | |
| 72 °C 10 min | 1× | Final elongation | |
| BMP4 and FGF3 | 94 °C 1 min | 1× | Initial denaturation |
| 98 °C 10 sec + 68 °C 9 min | 30× | Amplification | |
| 72 °C 10 min | 1× | Final elongation | |
Following the manufacturer’s instructions, amplicons were purified using the Agencourt AMPure XP PCR Purification systems (Beckman Coulter, Pasadena, CA). The purified PCR product was quantified using Qubit® 2.0 Fluorometer (Life Technologies, Carlsbad, CA, USA) and the Qubit® dsDNA HS Assay Kit. Before pooling, each amplicon (i.e., purified PCR product) from the 36 individuals was diluted to 1ng/µl. 5 µl of each amplicon from asingle participant was pooled and used as the starting material for library preparation.
Using the Nextera XT DNA library preparation kit (Illumina, Inc.), libraries were constructed using 1 ng of the pooled amplicons of each participant and indexed separately using IDT Illumina DNA/RNA UD indexes (Illumina, Inc.). The tagmentation, amplification, clean-up, libraryquality check, normalization, and library pooling stages are the primary steps of the Nextera XT DNA library preparation workflow. The libraries were finally pooled at afinal concentration of 2nM. PhiX was also included as asequencing control in the pool at aconcentration of 12.5 pM.
2nM of the pooled libraries were further subject to denaturation and dilution to aloading concentration of 10 pM for sequencing. Libraries were sequenced in Dr. D. Andrew Merriwether’s Molecular Anthropology Lab at Binghamton University. We used an Illumina MiSeq next-generation sequencer and MiSeq Reagent Kit v2 with a2×149-cycle paired-end run configuration (Illumina, Inc.). The reason for choosing a2×149 read length configuration rather than the commonly used ٢×١٥١ read length is that we used IDT for Illumina UD indexes, which are ١٠ bp long, as opposed to the Nextera XT indexes, which are 8 bp long.
The raw NGS output data was analyzed using the variant calling pipeline that utilizes various programs/tools including GATK, BWA, Picard, and Samtools (McKenna et al. 2010; DePristo et al. 2011; Van der Auwera et al. 2013; Poplin et al. 2018; Van der Auwera and O’Connor 2020). The variant calling pipeline uses aFASTQ file as astarting material which contains raw sequencing data with quality scores assigned to each base call (Cock et al. 2010). As the final output, the pipeline generates aVCF (Variant Call Format) file that contains the variants that were detected in the sample. The non-variablesections of thegenome, which comprise most of the human genome, are not included in the VCF file. Each step of the bioinformatics data analysis is provided in the Supplementary Materials. Alinkage disequilibrium analysis was conducted using LDlink software to distinguish between the causative variants and those in linkage disequilibrium (LD) (Machiela and Chanock 2015).
Spearman’s rank correlation coefficient (rho) was performed to estimate the relationship between adetected genetic variant on dental trait expression. In Spearman’s rank correlation tests, two ordered or ranked variables were correlated by their strength as well as their direction (Hauke and Kossowski 2011). We ranked the genotypes based on the number of copies of the alternate (A) and reference (R) alleles: 0: homozygotes for the reference allele (RR); 1: heterozygotes (RA); and 2: homozygotes for the alternate allele (AA) (Kimura et al. 2015). The other variable (i.e., level of dental trait expression) was already designed in the ranked format in the ASUDAS (Fig. 1) (Turner et al. 1991). In other words, degrees of expression of adental trait were classified on an ordinal scale starting with the lowest grade and ending with the highest grade.

Fig. 1. ASUDAS plaques illustrating the range of Carabelli’s cusp and shoveling expression. UM – Upper molar, UI 1 – Upper lateral incisor
Table 3 displays the prevalence of Carabelli’s trait and shoveling for each grade among the study participants. Three individuals exhibited grade 3, and four grade 4 Carabelli’s cusp expression. Atotal of 13 participants displayed the cusp form of Carabelli’s feature, which is grades 5 and above. Out of 36 participants, 16 displayed no Carabelli’s cups expression. On the other hand, most study participants demonstrated aweak expression of shoveling (i.e., grades 1 and 2) in UI1L and UI2L. In UI2L, one person demonstrated grade 3 shoveling, while another demonstrated grade 5. 21 and 19 individuals showed no shoveling in UI1L and UI2L, respectively.
| Trait | Grade | ||||||||
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | Total | |
| Carabelli’s cusp (UM1L) | 16 | 0 | 0 | 3 | 4 | 7 | 1 | 5 | 36 |
| Shoveling (UI1L) | 21 | 6 | 9 | 0 | 0 | 0 | 0 | 0 | 36 |
| Shoveling (UI2L) | 19 | 7 | 8 | 1 | 0 | 1 | 0 | 0 | 36 |
The variant calling pipeline identified 277 variants out of 48,665 bp DNA sequence that was amplified and sequenced in this study (see Tab. 1 for the amplicon lengths). The remaining 48,388 bp were excluded from the further statistical analysis since they did not exhibit any variable areas.
Among 277 variations, the Spearman’s rank correlation test showed moderate significant correlation between fifteen loci in four genes (WNT10A, SOSTDC1, FGF4, and FGF3) and the two dental phenotype (i.e., shoveling and Carabelli’s cusp) (Tab. 4). Significant negative and positive correlation was found between the dental features and two loci in WNT10A, seven loci in SOSTDC1, one locus in FGF4, and five loci in FGF3. The positions of the variations that were significantly linked with the dental features are shown in Figure2. BMP4 and NKX2-3 were excluded from the statistical analysis since the variant calling method did not identify any variation in these genes.
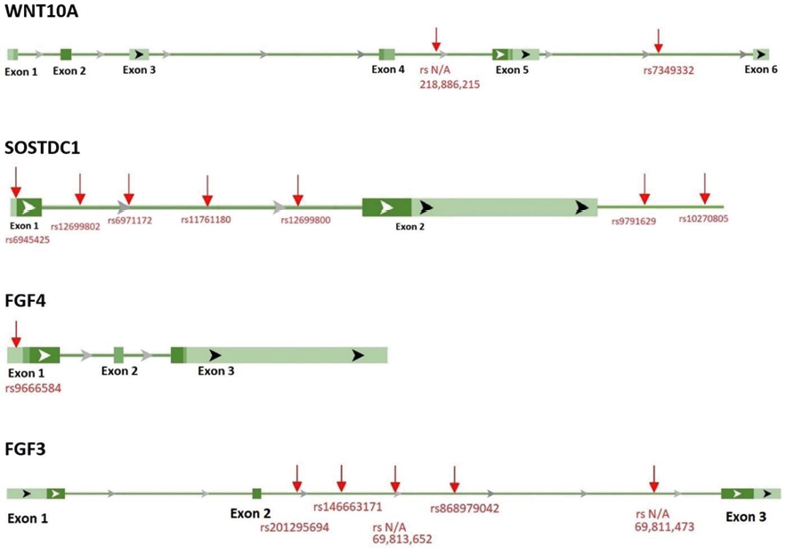
Fig. 2. Locations of the detected variants (Red arrows). Dark green bars indicate coding exons, and the light green bars indicate non-coding untranslated regions (UTRs). Horizontal lines that connect exons show the introns. White and black arrows indicate the direction of transcription from 5’ to 3’. Grey arrows in the introns indicate the direction of splicing. Green bars and connecting lines showing exon and intron regions were taken from the National Center for Biotechnology Information’s website (https://www.ncbi.nlm.nih.gov/)
The majority of the variations found in this study—11—were intron variants, as shown in Table 5. Two of the observed variants were 5 prime UTR variants, and two are downstream gene variants. Three of the fifteen mutations were INDELs, whereas the other twelve are SNPs.
Three of the found variants were INDELs, located at the following positions: chr2:218886215, chr11:69811473, and chr11:69813652. The reference and alternate alleles found by the variant calling pipeline in this study did not match the reference and alternate alleles in the NCBI’s dbSNP database (https://www.ncbi.nlm.nih.gov/snp/). In addition, no rs ID was assigned to the variant at chr11:69813652 by the variant calling pipeline. Therefore, the genomic positions (i.e., chr2: 218886215, chr11: 69811473, and chr11: 69813652) were used throughout the text instead of the rs numbers to avoid any confusion.
| Chromosome Position | SNP/INDEL ID | Gene | Rho | Sig. (2-tailed) |
N | Associated trait(s) |
| chr2:218891661 | rs7349332 | WNT10A | -0.437 | 0.026 | 26 | Shoveling UI2L |
| chr2: 218886215 | N/A | WNT10A | -0.458 | 0.019 | 26 | Carabelli’s cusp UM1L |
| chr7:16460517 | rs10270805 | SOSTDC1 | 0.378 | 0.023 | 36 | Shoveling UI1L |
| 0.430* | 0.009 | 36 | Shoveling UI2L | |||
| chr7:16460784 | rs9791629 | SOSTDC1 | 0.378 | 0.023 | 36 | Shoveling UI1L |
| 0.430* | 0.009 | 36 | Shoveling UI2L | |||
| chr7:16463435 | rs12699800 | SOSTDC1 | -0.329 | 0.050 | 36 | Carabelli’s cusp UM1L |
| chr7:16464081 | rs11761180 | SOSTDC1 | -0.329 | 0.050 | 36 | Carabelli’s cusp UM1L |
| chr7:16464648 | rs6971172 | SOSTDC1 | 0.340 | 0.043 | 36 | Shoveling UI1L |
| 0.388 | 0.019 | 36 | Shoveling UI2L | |||
| -0.366 | 0.028 | 36 | Carabelli’s cusp UM1L | |||
| chr7:16464881 | rs12699802 | SOSTDC1 | -0.329 | 0.050 | 36 | Carabelli’s cusp UM1L |
| chr7:16465716 | rs6945425 | SOSTDC1 | 0.416 | 0.012 | 36 | Shoveling UI1L |
| 0.341 | 0.042 | 36 | Shoveling UI2L | |||
| chr11:69775191 | rs9666584 | FGF4 | 0.450* | 0.006 | 36 | Shoveling UI2L |
| chr11:69813640 | rs868979042 | FGF3 | 0.354 | 0.034 | 36 | Carabelli’s cusp UM1L |
| chr11:69813824 | rs146663171 | FGF3 | 0.342 | 0.041 | 36 | Shoveling UI1L |
| chr11:69813844 | rs201295694 | FGF3 | 0.437* | 0.008 | 36 | Shoveling UI1L |
| 0.412 | 0.013 | 36 | Shoveling UI2L | |||
| chr11:69811473 | N/A | FGF3 | -0.349 | 0.037 | 36 | Carabelli’s cusp UM1L |
| chr11:69813652 | N/A | FGF3 | 0.405 | 0.014 | 36 | Carabelli’s cusp UM1L |
| Genomic Position | Variant ID | Variant type | Reference allele | Alternate allele | Gene | Consequence |
| chr2:218891661 | rs7349332 | SNP | C | T | WNT10A | intron variant |
| chr2:218886215 | N/A | INDEL | GCA | G | WNT10A | intron variant |
| chr7:16460517 | rs10270805 | SNP | A | G | SOSTDC1 | downstream gene variant |
| chr7:16460784 | rs9791629 | SNP | C | T | SOSTDC1 | downstream gene variant |
| chr7:16463435 | rs12699800 | SNP | C | T | SOSTDC1 | intron variant |
| chr7:16464081 | rs11761180 | SNP | A | G | SOSTDC1 | intron variant |
| chr7:16464648 | rs6971172 | SNP | G | C | SOSTDC1 | intron variant |
| chr7:16464881 | rs12699802 | SNP | T | C | SOSTDC1 | intron variant |
| chr7:16465716 | rs6945425 | SNP | T | C | SOSTDC1 | 5 prime UTR variant |
| chr11:69775191 | rs9666584 | SNP | A | G | FGF4 | 5 prime UTR variant |
| chr11:69813640 | rs868979042 | SNP | G | *,A | FGF3 | intron variant |
| chr11:69813824 | rs146663171 | SNP | G | *,A | FGF3 | intron variant |
| chr11:69813844 | rs201295694 | SNP | T | A,* | FGF3 | intron variant |
| chr11:69811473 | N/A | INDEL | GA | G | FGF3 | intron variant |
| chr11:69813652 | N/A | INDEL | ATGGATGGATGGGTGGATGGC | *,A | FGF3 | intron variant |
The LD heatmaps for the SOSTDC1, FGF3, and FGF4 variations are shown in Figures 3 and 4. The LD r2 and D’ values are provided in tables S1, S2, S3, and S4 in the Supplementary Materials. The degree of the LD between the variants is shown by the color intensity in the Figures 3 and 4. For each pair of variants, the r2 measurements are displayed in red, while the D’ measurements are displayed in blue. Due to the issues with inconsistent rs numbers, alternate alleles, and reference alleles encountered during data analysis, the variations at chr2:218886215, chr11:69811473, and chr11:69813652 were excluded from the LD analysis.
The r2 values for the SOSTDC1 variants show arange of correlations from “strong” to “weak” and “no LD” (Fig. 3). D’ values, on the other hand, exhibit no variation and all show asignificant correlation among all SOSTDC1 variants. Similarly, r2 values for the FGF3 and FGF4 variants all show no correlation between the variants in these genes. D’, on the other hand, indicates strong LD for many variants except for the associations between rs9666584, rs201295694, and rs146663171, which are depicted with alighter shade of blue (Fig. 4).
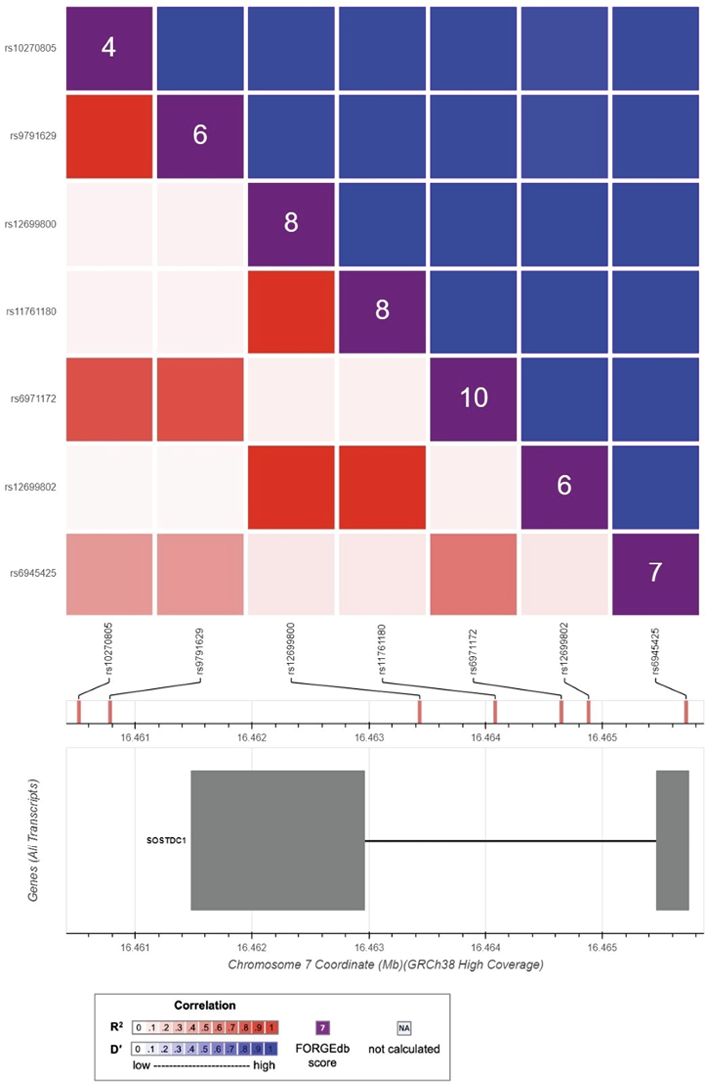
Fig. 3. Linkage disequilibrium heatmap showing correlations between SOSTDC1 variants
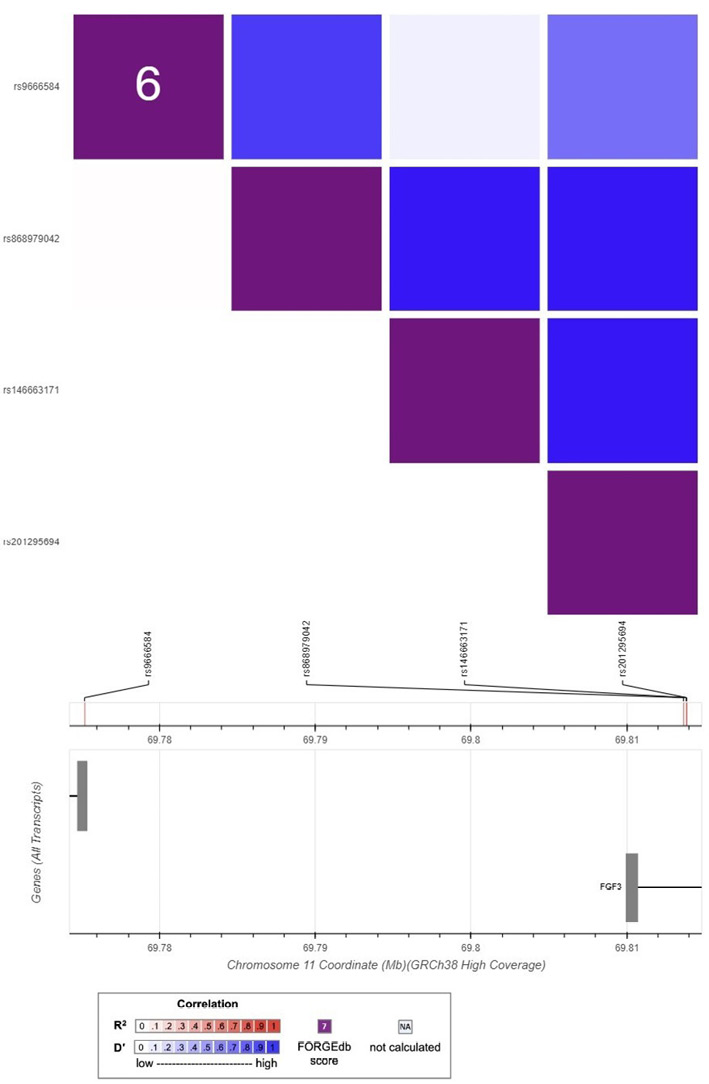
Fig. 4. Linkage disequilibrium heatmap showing correlations between FGF3 and FGF4 variants
The genetic pathways that are involved in the embryonic development of teeth have been described in detail over the past few decades. Hundreds of genes that ultimately produce teeth have been identified, and the gene network interactions that cascade during embryonic development to produce teeth are well understood (Pillas et al. 2010). However, only few genes that play asignificant role in the production of common dental nonmetric variation have been identified so far (Kimura et al. 2009, 2015; Lee et al. 2012). The etiology behind many nonmetric dental traits still remains to be elucidated.
The purpose of this study was to determine whether variation in the basal DNA sequences of six candidate genes, NKX2-3, SOSTDC1, BMP4, FGF3, FGF4, and WNT10A, which have been previously implicated in dental development, has impact on an individual’s expression of shovel-shaped incisors (SSI) and/or Carabelli’s cusp (CC). The findings reveal that fifteen SNPs/INDELs in SOSTDC1, FGF3, FGF4, and WNT10A significantly correlate with shoveling and/or the Carabelli’s cusp grades.
SSI and CC were negatively correlated with seven variants, including rs7349332 and chr2:218886215 in WNT10A; rs12699800, rs11761180, rs6971172, and rs12699802 in SOSTDC1; and chr11:69811473 in FGF3 (Tab. 4). The rho values vary from -0.329 to -0.458 which indicate amoderate negative association. Anegative Spearman’s rho value means that an individual having an increased number of the alternate allele(s) is correlated with adecreased level of trait expression. Six of the seven negatively correlated variants were associated with CC, and one is associated with SSI (UI2L). This finding is interesting because Carabelli’s cusp expression consistently displays negative associations with the alternate alleles in these six loci. In other words, at these loci, the reference allele is more linked to higher grades of Carabelli’s cusp expression compared to the alternate allele. Future research, including abigger sample size and amore geographically diversified group of study participants, is required to confirm these negative correlations.
SSI and CC expression was positively linked to nine variants: rs10270805, rs9791629, rs6971172, rs6945425 in SOSTDC1, rs9666584 in FGF4, rs868979042, rs146663171, rs201295694, and chr11:69813652 in FGF3 (Tab. 4). The rho values ranged between 0.340 and 0.450 indicating moderate correlations. Apositive Spearman’s value suggests that the higher grades of SSI and CC are associated with the number of alternate alleles.
Five SNPs, rs10270805, rs9791629, rs6971172, rs6945425, and rs201295694, were positively correlated with shoveling in both the central (UI1L) and lateral (UI2L) incisors (Tab. 4). These results are not unexpected given the previous research that showed that shoveling expression in central and lateral incisors is significantly correlated with each other (Hasegawa et al. 2009). In other words, if the upper central incisors in an individual exhibits shoveling, there is ahigh likelihood that the lateral incisors will also have some degree of shoveling expression. These observations can be explained by the fact that the same genetic factors affect both the central and lateral incisors (Hasegawa et al. 2009). The only variant that we found to be associated with both SSI and CC is the SNP rs6971172 in SOSTDC1. This particular SNP exhibited apositive correlation with SSI and anegative correlation with CC. This outcome is also not surprising, given that Carabelli’s cusp and shoveling are typically thought to have opposite manifestations across Asian-Native American and European populations (Scott et al. 2018). According to Scott et al. (2018), individuals from these regions often exhibit either shoveling or Carabelli’s cusp, but not (in most cases) both.
This study found no significant correlations between the NKX2-3 and BMP4 variants and the two dental traits of interest. However, it is important to note that this result does not necessarily meanthat these genes do not have any specific functions related to the formation of shoveling and Carabelli’s cusp. The lack of correlation may be attributed to the limited sample size used in this study. Thus, future research with alarger sample size is necessary to reliably determine the role of these genes in the formation of dental traits.
It is noteworthy that the majority of the variations detected in this study have not been reported before, but are included in the ClinVar database. ClinVar is apublicly available database that archives genetic variations with associated phenotypes or diseases (https://www.ncbi.nlm.nih.gov/clinvar/). The fact that the variants detected in this study have not been reported in scientific publications and yet are included in the ClinVar database may be due to the possibility that these SNPs/INDELs may have been discovered in earlier Genome-Wide Association Studies (GWAS), and their functions are still unknown. One exception to this is SNP rs7349332 in the WNT10A gene, which has been previously linked to increased risk of colorectal cancer, hair loss and hair shape (Li et al. 2017). Kimura et al. (2015) discovered asignificant correlation between crown size and rs7349332. In addition, Eriksson and colleagues (2010) reported an association between the alternate allele of this particular SNP and hair curl. In our study, this SNP was negatively associated with shoveling (UI2L) indicating that the increased number of alternate alleles at this locus is associated with decreased grade of shoveling expression in UI2L.
Introns make up the majority of the variants found in this study (Tab. 5). Figure 2 shows the visual representation of the locations of the discovered variants. Introns refer to non-coding sections of DNA, which do not provide instructions for the production of amino acids. Introns are often regarded as functionless DNA “junk” because they are removed during transcription and not involved in protein synthesis (Parenteau and Abou Elela 2019:923). Recently, crucial roles that introns play during gene expression have been recognized (Jo and Choi 2015). Immediately after transcription, introns are removed from the pre-mRNA transcript through aprocess known as splicing. Subsequently, the exons are joined together to form amature mRNA, which will eventually result in the production of the protein. The process of splicing does not always follow the same pattern. Although it is atightly controlled molecular mechanism, splicing errors and the alternative splicing events frequently occur which results in the alternative forms of aprotein from the same mRNA template. Alternative splicing is awidespread phenomenon, occurring in about 95% of multi-exon genes in humans (Jo and Choi 2015).
Contrary to the common belief, introns are evolutionarily advantageous as they are amajor source of novel genes and gene products (Jo and Choi 2015). The process of alternative splicing can potentially generate many isoforms of proteins from asingle gene. In addition, research revealed that genes with introns exhibit enhanced levels of gene expression than genes without introns (Shabalina et al. 2010).
The intron variants discovered in this study are unlikely to change the protein’s amino acid sequences or functions. Nonetheless, there is apossibility that these variants could modify gene expression through processes such as alternative splicing and gene expression enhancement. This highlights the necessity of performing deep sequencing of the entire gene region, encompassing both intronic and exonic regions, instead of focusing solely on exons. Variation in introns, the 3’ and 5’ untranslated regions (UTRs), and the level of gene expression can all affect the dental phenotypes such as the ones that are investigated in this study.
The results of the LD analysis suggest that there is significant linkage disequilibrium among multiple variants, which indicates anon-random association (Figures 3 and 4, and Table S1, S2, S3, and S4 in Supplementary Materials). This result suggests that the variants detected in this study may not actually be causing SSI and CC. Instead, they might be potentially linked to an as-of-yet unelucidated variant or variants that are nearby and under selection pressure. If that is the case, this makes SSI and CC “hitchhiking” phenotypes that are inherited together with another phenotype which might be the actual target of natural selection (Park et al. 2012:508). Therefore, the identification of the causal genes is made more difficult by the fact that the variations are in substantial LD. As aresult, it is unclear which SNPs or INDELs are actually responsible for SSI and CC. Future research needs to clarify in more detail how these genes interact with other genes and how they affect the expression of Carabelli’s cusp and shovel-shaping.
Supplementary materials available on the request.
The current study aimed to shed light on the genetic basis of Carabelli’s cusp and shovel-shaped incisors. The degree of expression of the two dental characteristics and fifteen short variations in FGF3, FGF4, SOSTDC1, and WNT10A were shown to be significantly correlated. The conclusion may be formed in part because of the small sample size that variations in the genes FGF3, FGF4, SOSTDC1, and WNT10A may play arole in the development of shoveling and Carabelli’s cusp. More research is required to determine if the genes NKX2-3 and BMP4 contribute to the development of shoveling and Carabelli’s cusp.
Acknowledgements
This study was funded by Binghamton University’s Transdisciplinary Areas of Excellence Program.
Conflict of interests
The authors declare no conflict of interests.
Authors’ contribution
FNE – conception and design of the study, funding acquisition, data collection, performing the lab work and data analysis, manuscript writing; DAM – conception and design of the study, funding acquisition, reviewing and editing, critical revision of the article. All authors have read and agreed to the published version of the manuscript.
Informed consent
The study protocol used in this project was approved by the Binghamton University Institutional Review Board (IRB). All participants volunteered in this study provided an informed consent form which was approved by the IRB at Binghamton University (STUDY00002536). Please visit https://www.binghamton.edu/research/division-offices/research-compliance/human-subjects/about.html#Contact%20Information for more information about the Binghamton University IRB office.
This article is based on Fatma Nur Erbil’s Ph.D. dissertation titled “GENETIC DETERMINANTS OF SHOVEL-SHAPED INCISORS AND CARABELLLI’S CUSP”. Please visit https://www.proquest.com/docview/2694486327?pq-origsite=gscholar&fromopenview=true&sourcetype=Dissertations%20&%20Theses for acopy of the dissertation.
Brook AH, Jernvall J, Smith RN, Hughes TE, Townsend GC. 2014. The dentition: the outcomes of morphogenesis leading to variations of tooth number, size and shape. Aust Dent J 59:131–142. https://doi.org/10.1111/adj.12160
Carayon D, Adhikari K, Monsarrat P, Dumoncel J, Braga J, Duployer B, et al. 2019. Ageometric morphometric approach to the study of variation of shovel-shaped incisors. Am J Phys Anthropol 168(1):229–241. https://doi.org/10.1002/ajpa.23709
Cock PJA, Fields CJ, Goto N, Heuer ML, Rice PM. 2010. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res 38(6):1767–1771. https://doi.org/10.1093/nar/gkp1137
DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. 2011. Aframework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43:491–498. https://doi.org/10.1038/ng.806
Eriksson N, Macpherson JM, Tung JY, Hon LS, Naughton B, Saxonov S, et al. 2010. Web-based, participant-priven studies yield novel genetic associations for common traits.PLoS genet6(6):e1000993. https://doi.org/10.1371/journal.pgen.1000993
Hanihara T. 2008. Morphological variation of major human populations based on nonmetric dental traits.Am J Phys Anthropol136(2):169–182. https://doi.org/10.1002/ajpa.20792
Hasegawa Y, Terada K, Kageyama I, Tsukada S, Uzuka S, Nakahara R, et al. 2009. Influence of shovel-shaped incisors on the dental arch crowding in Mongolian females. Okajimas Folia Anat Jpn 86(2):67–72. https://doi.org/10.2535/ofaj.86.67
Hauke J, Kossowski T. 2011. Comparison of Values of Pearson’s And Spearman’s Correlation Coefficients on the Same Sets of Data.Quaest Geogr٣٠(٢):87–93. https://doi.org/10.2478/v10117-011-0021-1
Hlusko LJ, Carlson JP, Chaplin G, Elias SA, Hoffecker JF, Huffman M, et al. 2018. Environmental selection during the last ice age on the mother-to-infant transmission of vitamin D and fatty acids through breast milk. PNAS 115(19):E4426–E4432. https://doi.org/10.1073/pnas.1711788115
Hrdlička A. 1920. Shovel-shaped teeth. Am J Phys Anthropol 3(4):429–465. https://doi.org/10.1002/ajpa.1330030403
Hughes TE, Townsend GC. 2013. Twin and family studies of human dental crown morphology: Genetic, epigenetic, and environmental determinants of the modern human dentition. In: GR Scott and JD Irish, editors. Anthropological Perspectives on Tooth Morphology: Genetics, Evolution, Variation. New York, United States: Cambridge University Press, 31–68.
Irish JD, Morez A, Flink LG, Phillips EL W, Scott GR. 2020. Do dental nonmetric traits actually work as proxies for neutral genomic data? Some answers from continental‐ and global‐level analyses.Am J Phys Anthropol172(3):347–375. https://doi.org/10.1002/ajpa.24052
Jernvall J, Thesleff I. 2000. Reiterative signaling and patterning during mammalian tooth morphogenesis. Mech Dev 92(1):19–29. https://doi.org/10.1016/s0925-4773(99)00322-6
Jo BS, Choi SS. 2015. Introns: The Functional Benefits of Introns in Genomes.Genomics Inform13(4):112–118. https://doi.org/10.5808/GI.2015.13.4.112
Jussila M, Thesleff I. 2012. Signaling Networks Regulating Tooth Organogenesis and Regeneration, and the Specification of Dental Mesenchymal and Epithelial Cell Lineages. Cold Spring Harb Perspect in Biol 4(4):a008425. https://doi.org/10.1101/cshperspect.a008425
Kimura R, Watanabe C, Kawaguchi A, Kim YI, Park SB, Maki K, et al. 2015. Common polymorphisms in WNT10A affect tooth morphology as well as hair shape. Hum Mol Genet 24(9):2673–2680. https://doi.org/10.1093/hmg/ddv014
Kimura R, Yamaguchi T, Takeda M, Kondo O, Toma T, Haneji K, et al. 2009. Acommon variation in EDAR is agenetic determinant of shovel-shaped incisors. Am J Hum Genet 85(4):528–535. https://doi.org/10.1016/j.ajhg.2009.09.006
Lee WC, Yamaguchi T, Watanabe C, Kawaguchi A, Takeda M, Kim YI, et al. 2012. Association of common PAX9 variants with permanent tooth size variation in non-syndromic East Asian populations. J Hum Genet 57(10):654–659. https://doi.org/10.1038/jhg.2012.90
Li P, Liu W, Xu Q, Wang C. 2017. Clinical significance and biological role of Wnt10a in ovarian cancer.Oncol Lett14(6):6611–6617. https://doi.org/10.3892/ol.2017.7062
Machiela MJ, Chanock SJ. 2015. LDlink: aweb-based application for exploring population- specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics 31(21):3555–3557. https://doi.org/10.1093/bioinformatics/btv402
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. 2010. The Genome Analysis Toolkit: aMapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20(9):1297–1303. https://doi.org/10.1101/gr.107524.110
McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F. 2010. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor.Bioinformatics 26(16):2069–2070. https://doi.org/10.1093/bioinformatics/btq330
Parenteau J, Abou Elela S. 2019. Introns: good day junk is bad day treasure.Trends Genet35(12):923–934. https://doi.org/10.1016/j.tig.2019.09.010
Park JH, Yamaguchi T, Watanabe C, Kawaguchi A, Haneji K, Takeda M, et al. 2012. Effects of an Asian-specific nonsynonymous EDAR variant on multiple dental traits. J Hum Genet 57(8):508–514. https://doi.org/10.1038/jhg.2012.60
Pillas D, Hoggart CJ, Evans DM, O’Reilly PF, Sipilä K, Lähdesmäki R, et al. 2010. Genome-Wide Association Study Reveals Multiple Loci Associated with Primary Tooth Development during Infancy. PLoS Genet 6(2):e1000856. https://doi.org/10.1371/journal.pgen.1000856
Poplin R, Ruano-Rubio V, DePristo MA, Fennell TJ, Carneiro MO, Van der Auwera GA, et al. 2018. Scaling accurate genetic variant discovery to tens of thousands of samples. bioRxiv 1–22. https://doi.org/10.1101/201178
Scott GR. 1980. Population variation of Carabelli’s trait. Hum Biol 52(1):63–78. Available through: JSTOR website https://www.jstor.org/stable/41463185 [Accessed 7 June 2024].
Scott GR, Irish JD. 2017. Human tooth crown and root morphology: The Arizona State University Dental Anthropology System. Cambridge University Press.
Scott GR, Turner II CG, Townsend GC, Martinón-Torres M. 2018. The Anthropology of Modern Human Teeth: Dental Morphology and its Variation in Recent and Fossil Homo sapiens. 2nd edition. Cambridge University Press. https://doi.org/10.1017/9781316795859
Shabalina SA, Ogurtsov AY, Spiridonov AN, Novichkov PS, Spiridonov NA, Koonin EV. 2010. Distinct patterns of expression and evolution of intronless and intron-containing mammalian genes.Mol Biol and Evol27(8):1745–1749. https://doi.org/10.1093/molbev/msq0864
Tabor HK, Risch NJ, Myers RM. 2002. Candidate-gene approaches for studying complex genetic traits: practical considerations. Nat Rev Genet 3:391–397. https://doi.org/10.1038/nrg796
Thesleff I. 2006. The genetic basis of tooth development and dental defects. Am J Med Genet Part A140A:2530–2535. https://doi.org/10.1002/ajmg.a.31360
Tsai PL, Hsu JW, Lin LM, Liu KM. 1996. Logistic analysis of the effects of shovel trait on Carabelli’s trait in a Mongoloid population. Am J Phys Anthropol 100(4):523–530. https://doi.org/10.1002/(SICI)1096-8644(199608)100:4<523::AID-AJPA6>3.0.CO;2-R
Turner CG, Nichol CR, Scott GR. 1991. Scoring procedures for key morphological traits of the permanent dentition: the Arizona State University Dental Anthropology system. Advances in Dental Anthropology, New York: Wiley Liss, 13–31.
Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, del Angel G, Levy-Moonshine A, et al. 2013. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit Best Practices Pipeline.Curr Protoc Bioinform43(1):11.10.1–11.10.33. https://doi.org/10.1002/0471250953.bi1110s43
Van der Auwera, GA, O’Connor BD. 2020. Genomics in the Cloud: using Docker, GATK, and WDL in Terra (1st edition), O’Reilly Media.
Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. 2012. Primer-BLAST: Atool to design target-specific primers for polymerase chain reaction. BMC Bioinform 13,134:1–11. https://doi.org/10.1186/1471-2105-13-134

